A spatial control for correct timing of gene expression during the Escherichia coli cell cycle
Yuan Yao1,¶, Lifei Fan1,¶,,Yixin Shi1,2,Ingvild Odsbu3 and Morigen1, *
1School of Life Sciences, Inner Mongolia University, Hohhot 010021, China
2School of Life Sciences, Arizona State University, Temple, Arizona 85287-4501, USA
3Department of Public Health Sciences, Karolinska Institutet, Stockholm, Sweden
* Correspondence: morigenm@hotmail.com; Tel.: +86-471-4992442
¶These authors equally contributed to this work.
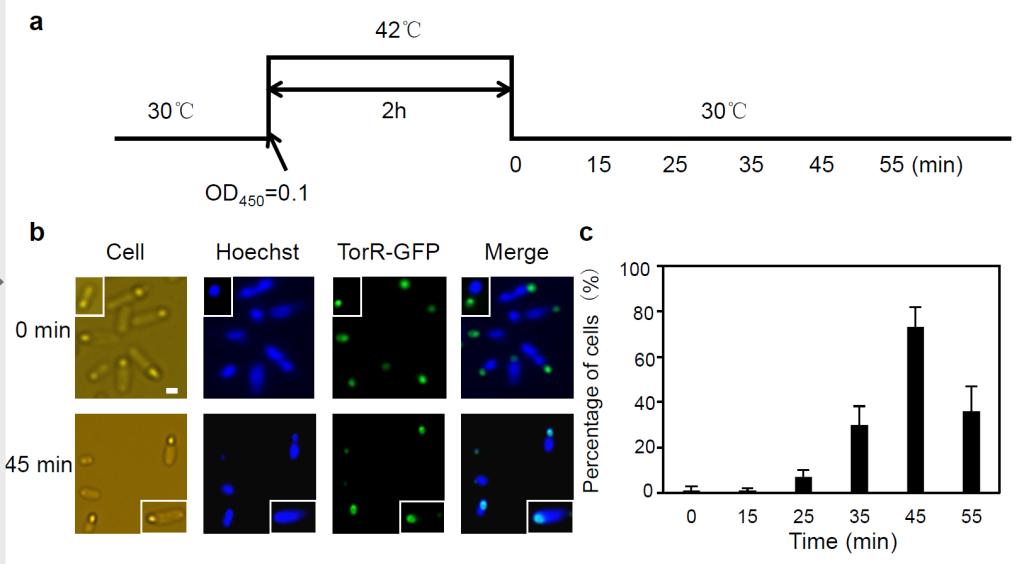
Temporal transcriptions of genes are achieved by different mechanisms such as dynamic interaction of activator and repressor proteins with promoters, and accumulation and/or degradation of key regulators as a function of cell cycle. We find that the TorR protein localizes to the old poles of the Escherichia coli cells, forming a functional focus. The TorR focus co-localizes with the nucleoid in a cell-cycle-dependent manner, and consequently regulates transcription of a number of genes. Formation of one TorR focus at the old poles of cells requires interaction with the MreB and DnaK proteins, and ATP, suggesting that TorR delivering requires cytoskeleton organization and ATP. Further, absence of the protein-protein interactions and ATP lead to loss in function of TorR as a transcription factor. We propose a mechanism for timing of cell-cycle-dependent gene transcription, where a transcription factor interacts with its target genes during a specific period of the cell cycle by limiting its own spatial distribution.
Original URL: http://www.mdpi.com/2073-4425/8/1/1